
In vitro modelling of the mucosa of the oesophagus and upper digestive tract: narrative review
Introduction
The human ‘swallowing apparatus’ consists of the pharynx, the upper oesophageal sphincter (UOS), the oesophagus, and the lower oesophageal sphincter (LOS). Major functions of the oesophagus are to co-ordinate the movement of food from the mouth to the stomach, and to control the retrograde movement of digestive material and digestive secretions—which can become pathological if excessive (1). While ingested food transits through the oesophagus in a relatively short period of time compared to distal parts of the gastrointestinal tract (GIT), oesophageal dysfunction appears to result in negative consequence on both health and quality of life. Disorders of the oesophagus include heartburn, dysphagia, eosinophilic oesophagitis, achalasia, oesophageal spasm, gastroesophageal reflux disease (2).
Several methodologies for in vitro simulation of the digestive tract are available to researchers, however, few of these consider the role of the oesophagus. This is part of a wider limitation of model gut systems in accurately modelling the digestive mucosa from both an anatomical and functional perspective.
Current model gut systems have been reviewed extensively elsewhere with reviews variously describing in vitro models by:
- Type; static models (3), dynamic models (4), cell based models (5-7), ex vivo models (8);
- Functionality; models of microbiome (9), physiological relevance (10,11);
- Or application; food digestion (12), drug delivery (13), screening and toxicity (14), assessment of pre- and pro-biotics (15), protein stability (16), nutraceutics (17).
These systems have many benefits including controllability, repeatability, ethics, timing and cost (7), but known limitations include modelling of hormonal control, simulation of physical forces [“the topography, motility and flow” (7)], immune function of the digestive tract, and accurate modelling of the absorptive mucosa.
Accurate modelling of the mucosa is an aspect of gastrointestinal physiology absent from the vast majority of these models and has only been the focus of limited discussion in the literature. Here, in a review of current literature, we aim to address this notable omission, discussing models of the oesophageal mucosa and the digestive and absorptive mucosa of the upper GIT including mucus function, and integration of mucus modelling in to in vitro digestion and absorption systems.
We present the following article in accordance with the Narrative Review reporting checklist (available at https://aoe.amegroups.com/article/view/10.21037/aoe-20-90/rc).
Static and dynamic digestion models
In vitro digestion models aim to simulate the processes of digestion and absorption carried out by the organs of the GIT (Table 1). Static digestion models have been reviewed extensively by Alegria et al. (3), and in an effort to harmonise the various static methodologies, the COST INFOGEST partnership developed a consensus method for static digestion modelling with a view to facilitating comparison of data and repeatability (18).
Table 1
| Organ | Function |
|---|---|
| Mouth (oral phase) | Breaks down foods into small particles via mastication |
| Lubrication of food for bolus formation and swallowing via saliva | |
| Digestive enzymes in saliva (amylase and lingual lipase) begin digestion | |
| Oesophagus | Transport of ingested material from the mouth to the stomach |
| Sphincters in the lower and upper oesophagus prevent reflux of stomach contents into the mouth and airways, while still allowing regurgitation and vomiting when required | |
| Stomach (gastric phase) | Storage of food |
| Chemical digestion via hydrochloric acid and pepsin | |
| Mechanical digestion via churning | |
| Pathogen defence via strong acid, and pepsin | |
| Duodenum (SI phase) | Neutralises stomach acids with bicarbonate secretions |
| Mixes chyme with bile from the liver for lipid digestion | |
| Mixes chyme with pancreatic enzymes for carbohydrate, protein, and lipid digestion | |
| Jejunum (SI phase) | Bulk absorption of simple carbohydrates, peptides, amino acids, and fatty acids |
| Ileum (SI phase) | Absorbs nutrients not taken up by the jejunum, e.g., vitamin B12 |
| Also important for uptake and redistribution of bile salts and remaining water-soluble vitamins | |
| Large intestine (colonic phase) | Reabsorbing water and vitamins not absorbed by small intestine |
| Housing the large intestinal microbiome | |
| Absorbing SCFAs produced by the large intestinal microbiome | |
| Storage of faeces |
Static models simulate luminal aspects of digestion in a stepwise manner, modelling the chemical and enzymatic environment of successive organs of the digestive tract (summarised in Tables 2-4) without taking into account some of the more complex physiological processes of the digestive tract that dynamic models aim to replicate including; secretion, gradual pH changes, physical forces of mixing, shearing and motility, gastric emptying, mucus permeation, absorption, and hormonal control.
Table 2
| Region | Name | Secretion | pH optima | Function |
|---|---|---|---|---|
| Mouth | Lingual lipase | Von Ebner glands (19) | 4.5–5.5 (19) | Digestion of triglyceride aggregates via ester bond hydrolysis, digestion continues until material reaches the stomach (19) |
| Salivary amylase | Parotid gland (20) | 5.6–6.9 (21) | Digestion of insoluble starches into progressively smaller soluble starches, with smallest being maltose, via hydrolysis of 1,4 glycosidic bonds (20) | |
| Stomach | Pepsin | As pepsinogen from chief cells (22) | 2 (22,23) | Pepsinogen autolysed into pepsin. Pepsin digests proteins via hydrolysis of peptide bonds. Pepsis cleaves at the C-terminal of hydrophobic, preferentially aromatic, i.e., phenylalanine, tyrosine and tryptophan, residues (22,23) |
| Gastric lipase | Chief cells (24) | 4.5–5.5 (25) | Similar to lingual lipase, triglycerides are digested via hydrolysis of ester bonds linking fatty acids to glycerol (25) | |
| Hydrochloric acid | Parietal cells (22) | – | Gastric HCl converts pepsinogen into pepsin rendering it an active protease, the acid also acts corrosively on proteins causing them to denature, exposing the peptide bonds to pepsin (12) | |
| Small intestine | Trypsin | Acinar cells as trypsinogen (26) | 7.0–8.0 (26) | Trypsinogen, the trypsin zymogen, is activated by enterokinase and/or trypsin via cleavage in the duodenum. Active trypsin then cleaves peptide bonds on the carboxy side of lysine and arginine (26) |
| Chymotrypsin | Acinar cells as chymotrypsinogen (27) | 7.0–8.0 (27) | Chymotrypsinogen, the chymotrypsin zymogen, is activated via cleavage by trypsin and autolysis. The active enzyme hydrolyses peptide bonds at the n-terminal side of aromatic amino acid residues (27) | |
| Carboxypeptidase A | Acinar cells as procarboxypeptidase (28) | ~7 (29) | Secreted as procarboxypeptidase and activated via trypsin cleavage, carboxypeptidase is an exopeptidase that cleaves peptide bonds of peptides with amino acids with free COOH groups giving a single amino acid as product. Carboxypeptidase A does not cleave peptide bonds when the relevant amino acid is proline, lysine, arginine or histidine (28) | |
| Elastases | Acinar cells (30) | 8.2–9.2 (31) | Elastases are serine proteases which cleave peptide bonds on the c-terminal side of hydrophobic amino acids (30) | |
| Pancreatic lipase | Acinar cells (32) | 8 (32) | Pancreatic lipase, following binding of its cofactor—colipase, hydrolyses ester bonds of triglycerides at the oil-water interface after the emulsification of lipids by bile. The resulting product is 2 free fatty acids and a monoacylglycerol (32) | |
| Phospholipases | Acinar cells (33) | 6–8 (34) | Phospholipase enzymes act by hydrolysing the ester bonds of phospholipids, releasing a fatty acid (33) | |
| Pancreatic amylase | Acinar cells (35) | 7 (36) | Like salivary amylase, pancreatic amylase hydrolyses 1,4 glycosidic bonds in oligosaccharides, resulting in maltose, maltotriose and limit dextrins (37) |
Table 3
| Class | Name | Function |
|---|---|---|
| Carbohydrate | Sucrase-isomaltase | Sucrase isomaltase functions as carbohydrase dimer of sucrase and isomaltase. The sucrase subunit hydrolyses the glycosidic bond in sucrose to release glucose and fructose. The isomaltase subunit hydrolyses 1,6 glycosidic bonds in limit dextrins (38) |
| Lactase | Hydrolyses the β-1,4 glycosidic bonds in lactose, found in milk and dairy products, into glucose and galactose (38) | |
| Maltase-glucoamylase | Composed of two subunits, maltase and glucoamylase, which with differing substrate specificity hydrolyse α-1,4 glycosidic bonds at the non-reducing side of oligosaccharides in the gut lumen, releasing terminal glucose (38) | |
| Trehalase | Hydrolyses the 1,1 glycosidic bonds found in trehalose, releasing glucose (38) | |
| Proteins | Peptidases | There is a vast array of peptidases found in the brush border of the small intestine, and these enzymes display varying degrees of substrate specificity, i.e., target amino acids and length of peptide, these specificities have been previously discussed (39) |
| Lipids | Phospholipase | Phospholipase A2 has been noted to be present in the brush border of the small intestine, where it will hydrolyse bonds which link fatty acids to phospholipids (40) |
Table 4
| Name | Secretion | Function |
|---|---|---|
| Cholecystokinin (CCK) | Duodenal enteroendocrine cells (41) | While Known for a role in satiety, CCK also stimulates the secretion of pancreatic enzymes and hormones and stimulates motility of the gastrointestinal tract. CCK secretion is stimulated by the presence of partially digested proteins and gastric acid in the duodenum (42) |
| Gastrin | G cells (43) | Gastrin, secreted from G cells in the stomach and duodenum, stimulates secretion of gastric acid, as well as pepsinogen and intrinsic factor from parietal cells, and secretin (42) from S cells (44) |
| Secretin | S cells (44) | Secretin, secreted by S cells in the duodenum, inhibits the release of gastrin and gastric acid, lowers pressure of the lower oesophageal sphincter, and stimulates pancreatic release of bicarbonates, digestive enzymes and insulin (44) |
| Vasoactive intestinal polypeptide (VIP) | Neurons and immune cells (45) | Secretion of water and electrolytes into the duodenum following the secretion of pancreatic juices and bile. Also inhibits gastric acid secretion and relaxes smooth muscles of the gut (42) |
| Gastric inhibitory peptide (GIP) | SI K cells (46) | Secreted primarily in the duodenum and upper jejunum GIP inhibits gastric secretions and stimulates secretion of somatostatin which intensifies the downregulation of gastric secretions (47) However, the most important role of GIP is the potent stimulation of insulin release (42) |
| Other peptides | Vary | There are other hormones and peptides that have an influence on gastrointestinal regulation, albeit to a lesser extent than those above. These include somatostatin which downregulates gastrointestinal processes, ghrelin which stimulates gastric secretions during hunger, motilin which stimulates gastrointestinal motility and many others (42) |
The functionalities of certain dynamic gastrointestinal models are summarised here, in brief. The Dynamic Gastric Model (DGM) (48), Human Gastric Simulator (HGS) (49) and the Human Gastric Digestion Simulator (HGDS) (50) are computer-controlled mono-compartmental systems (MoCS) that aim to recreate gastric secretions, churning and emptying.
The DGM consists of a flexible main body surrounded by a water jacket, a piston and barrel recreate peristaltic forces by fluctuating the water pressure in the jacket to simulate contractions in the fundus and antrum (48). The HGS, is a latex vessel subject to simulated gastric forces from mechanical, belt-driven rollers (49). Data from the DGM and HGS (51-53), is reported to correlate well with in vivo data and the systems have been used for modelling transit and emptying times, gastric enzymatic digestion, particle breakdown, rupture times of delivery systems, phase separation, gel formation and nutrient/drug release (48).
The TNO Gastro-Intestinal Model (TIM-1), The DIDGI MGS, and the Engineered Stomach and small INtestinal MGS (ESIN) are multi-compartmental models of the upper GIT. The TNO Gastro-Intestinal Model (TIM-1) consists of four compartments simulating: the stomach, duodenum, jejunum and ileum (54-57). The DIDGI MGS (58) simulates the upper GIT with a 2-compartment system, representing the stomach and small-intestine—both stirred glass vessels surrounded by a heated water jacket. The pH changes, secretion rates and emptying times are computer controlled, based on data collected from in vivo experiments. The ESIN (59) is comprised of 6 compartments, including: a meal reservoir (R1) which allows continuous passage of food material into the stomach compartment, a salivary ampoule (R2) which mixes the food components with salivary enzymes and buffers, the stomach (R3), the duodenum (R4), the jejunum (R5) and the ileum (R6).
While some of these systems have the capacity to model the oral phase and/or salivary secretions, none of them model oesophageal function. This reflects the limited role the oesophagus is considered to have in digestion, acting as a transport corridor between the mouth and stomach. However, modelling oesophageal function and interaction with the oral secretions and refluxed material from the upper digestive tract could bring important understanding of oesophageal pathologies.
Physiology of the oesophagus
The oesophagus is a 25–30 cm long, hollow, muscular tube, which connects the oral cavity and pharynx to the stomach (60). The oesophagus is split into two sections, the upper cervical section which makes up the first 2–6 cm of the oesophagus, and lower thoracic section (61). The UOS forms the junction between the pharynx and the cervical oesophageal region, and the LOS forms the junction between the thoracic oesophageal region and the stomach (60). The oesophagus transports ingested material, often in the form of a food bolus, from the oral cavity to the stomach via peristaltic waves which propagate down the oesophagus, shifting the bolus towards the stomach (62). A secondary function is to prevent reflux of gastric contents back into the oral cavity and to allow regurgitation and vomiting—a function mediated by the oesophageal sphincters (62).
Tissue structure
The oesophageal tissue is comprised of 4 layers: the mucosa, the submucosa, the muscularis externa and the adventitia (63). The mucosa is the uppermost layer on the luminal side of the oesophagus. It is composed of a surface epithelium (Figure 1A), a lamina propria and muscularis mucosae (Figure 1B). The oesophageal surface epithelium is a non-keratinised, stratified, squamous epithelia of around 30 cell layers (63-65). These cells are present throughout the oesophagus until a small area surrounding the LOS where gastrointestinal-like columnar cells may be observed (63). In Barrett’s Oesophagus, squamous epithelial cells around the LOS are replaced with columnar epithelial cells due to metaplasia (69). Found in the lower mucosal layers, below the basement membrane, is the lamina propria—a loose connective tissue rich with immune cells (63)—and the muscularis mucosae, a thin layer of longitudinal muscle cells (70). The mucosa is the primary defence against underlying tissue damage from acid refluxate (i.e., hydrochloric acid and pepsin), generating both pre-epithelial and epithelial defensive actions, most of which are provided by the specialised epithelium (65).
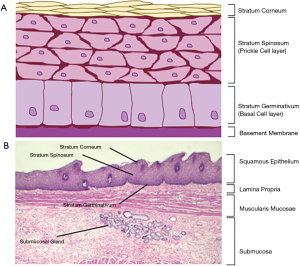
The submucosa (Figure 1B) is comprised of connective tissue, lymphocytes, plasma cells, the submucosal plexus and submucosal glands (63) which secrete bicarbonate ions and mucin (71). Bicarbonate ions function to neutralise acids during reflux of gastric contents into the oesophagus (71). Secreted mucin, MUC5B specifically in this case (72,73), typically forms surface mucus (74-76). In the oesophagus however, it has been noted that the levels of MUC5B secretion by submucosal glands are not sufficient to provide a fully functioning surface mucus layer (hence the absence of a mucus layer in the oesophagus) (77), thus the mucin secretions most likely function to help lubricate the bolus as it passes through the oesophagus.
The muscularis propria is important for the generation of peristaltic waves which propagate boluses down the oesophagus. It consists of up to 33% striated muscle in the cervical region and transitions to smooth muscle in the lower thoracic region. There is a transition region as the striated muscle begins to change to smooth muscle, this is identified by a mixture of striated and smooth muscle cells and a lack of significant contractions (62,63). Interestingly, striated muscle of the upper cervical region is not controlled voluntarily despite being innervated with somatic motor neurons, instead being controlled by several reflex inputs which bring about peristaltic contractions. On the contrary, smooth muscle present in the thoracic region of the oesophagus creates peristaltic waves via stimulation of the myenteric plexus (61).
Finally, and unlike the remainder of the GIT, the oesophagus does not contain a serosa layer, instead having an adventitia, a base layer of fixed connective tissues, binding the oesophagus to the surrounding tissue and holding it in place (63).
The oesophageal sphincters
The UOS sphincter is the musculo-cartilaginous high-pressure zone at the end of the pharynx and start of the cervical region of the oesophagus. This anatomical sphincter serves as the first barrier to the oesophagus. It prevents both the reflux of gastric contents into the oral cavity, of which hydrolytic enzymes and acid would cause damage to tissues, and the flow of inhaled air from entering the digestive tract, while also allowing vomiting and regurgitation in the necessary scenarios. Like the rest of the cervical portion of the oesophagus, the muscles surrounding the UOS are striated, skeletal muscles. Moreover, it is similarly controlled involuntarily, despite being under the influence of somatic nerves, being controlled rather on the basis of a variety of reflex inputs to the innervated motor neurons (70,78).
The LOS sphincter is not easily identified as the characteristic thickening of muscle tissue common to anatomical sphincters is not seen with the LOS as functionality occurs through maintenance of muscle tone, modulated by several excitatory and inhibitory vagal pathways in the muscles modulating sphincter function (70,79-83). The functionality is controlled by two muscles; the smooth muscle of the lower thoracic oesophagus, and striated muscles fibres found in the crural diaphragm (82,84). The smooth muscle is organised in C-shapes, rather than rings, which interact with gastric sling fibres. When in a contracted state, the smooth muscles clasp together and pull in the gastric sling fibres, creating a closure, and during relaxation, in the instance of passing food for example, the clasp is released to create an opening (85). The striated diaphragmatic sphincter muscles work in tandem with the smooth muscle providing functional support (81,85).
Oesophageal defence and associated disease
The surface mucus gel coating the epithelium and secreted bicarbonates form a pre-epithelial defence in the majority of the GIT (86,87). For example, mucin glycoproteins in gel format provide an exclusion barrier in the small intestine by forming pores around 200 nm in size, providing protection against larger toxic particles, enzymes and microorganisms (88). Moreover, mucus in the stomach and small intestine protects the epithelia from enzymatic degradation (86), while facilitating the production of an unstirred water layer, rich in secreted bicarbonate ions to neutralise the harsh acid present in the stomach and duodenum (87). Mucus layers in the GIT, particularly those formed in the intestine by MUC2, are also known to produce an exceptionally rigid, firm mucus layer close to the epithelia which is often sterile even in the colon—a region where bacterial counts are ×1010–×1012 per gram faecal matter—implicating its function as a mucosal protectant from microorganisms.
Though the oesophagus is associated with secreted mucus, with oesophageal submucosal glands strongly expressing and secreting mucin MUC5B (73), oesophageal epithelia scrapings have shown that the secretion is not significant enough to form a functioning surface mucus layer (0.47 µg/cm-2 oesophageal scrapings vs. 500 µg/cm-2 of stomach scrapings) (77). In the healthy oesophagus, bicarbonate secretions and salivary bicarbonates, are enough to limit epithelial damage to the oesophagus by acids (77). However, when acid levels are higher than normal during a reflux event the oesophagus is vulnerable to damage without a protective bicarbonate rich mucus layer (87).
Like the epithelium of the oral cavity, stratified squamous epithelium protects the delicate tissues below from abrasion, but the stratified squamous epithelia of the oesophagus have other specialised mechanisms that allow it to cope with acid reflux. The apical membrane has been shown to prevent H+ permeation into cells despite the presence of non-selective cation channels—it is thought that low pH causes a reduced influx of cations (39). Moreover, the apical junction complex, a complex which fuses adjacent cells in the stratum corneum and spinosum, regulates paracellular diffusion of H+ ions. There are 3 structures in total which make up the complex: closest to the apical side of the membrane are the tight junctions, this protein structure spans over the membranes of two cells and interlinks their cytoskeletons, bringing the membranes tight to one another. This complex is the main paracellular transport modulator of the complex. Found below the tight junctions are the adherens junction and found further below, close to the basolateral membrane, are desmosomes. Both protein structures provide structural support for the tight junction, while also having other regulatory roles within the cell. It is worth noting however that MUC1, a transmembrane mucin expressed in the surface epithelia of the oesophagus (73),may provide some protective functions, as studies have shown that MUC1 contains Lewis antigens—a feature of the epithelial glycocalyx where H. pylori is known to occur. In this circumstance, the MUC1 Lewis antigens act as a decoy by binding the H. pylori—this is followed by a complete release of MUC1 from the membrane, with MUC1 establishing its protective role by carrying the infective material far from the epithelia (89,90).
Despite these defence mechanisms, they are often easily overcome by unusually high levels of gastric reflux of acid and enzymes. High levels of reflux are often associated with disease, and depending on the severity of the cause, they can cause varying degrees of damage to the oesophageal mucosa, from minor issues, like tissue damage from acids, to serious issues like Barratt’s oesophagus and oesophageal adenocarcinoma (OADC).
In vitro oesophagus models
In vivo and ex vivo animal models, typically rodents like mice and rats, have been used to investigate oesophageal physiology (91,92). However, substantial differences between human and mouse oesophagus are a limitation. For example, the rodent oesophageal epithelia consists of around 4–6 layers, whereas that of a human consists of much thicker 25–30 layers and contains papillae like structures (91,92). Moreover, the mucosal layers of rodents are keratinised, as opposed to the non-keratinised cells present in humans, and do not contain mucus secreting glands (93). Oesophageal human cell models are widely used today (94) and continue to advance in complexity—with poor in vivo correlation of results from 2D monolayer methods leading to a focus on developing 3D models that accurately recapitulate the architecture and functionality of oesophageal tissue (95).
Organotypic 3D Raft Culture (OTC)
Transwell based Organotypic 3D Raft Culture (OTC) models utilise a transwell system in which submerged fibroblast cells, grafted onto a collagen matrix on the surface of the apical transwell membrane, act as a platform or “raft” for keratinocytes to differentiate into a functioning epithelial layer (Figure 2) (96,97). This model has been applied to a variety of tissues and have been used widely to investigate tissue development, disease and also for toxicity assays (97). OTC has been successfully used to model oesophageal epithelial generation and proliferation (98,99), oesophageal squamous cell carcinoma (ESCC) (100-102), eosinophilic oesophagitis (94) and Barratt’s oesophagus associated OADC (103-105).
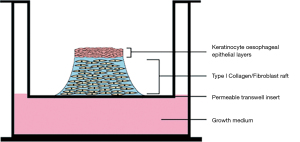
For example, in the work of Kalabis et al. [2012] (96), oesophageal fibroblasts were embedded in bovine collagen type I on an insert on top of a collagen matrix coated membrane and cultured for 7 days, forming a collagen/fibroblast raft on the insert membrane. Oesophageal keratinocytes were then seeded on the surface of the matrix and allowed to develop over the course of 4 days.Keratinocytes present at the air/liquid interface promote stratification and differentiation in the epithelial layer, generating epithelial layers representative of the oesophageal mucosa, recreating the stratum corneum, stratum spinosum and stratum germinativum, though the underlying tissue structures—which remain important to oesophageal function—are not recreated using this system.
3D multicellular spheroid culture and sphere formation assays
Multicellular spheroid culture, the first of the submerged 3D models used in oesophageal research, allows dissociated epithelial cells to reaggregate under submerged conditions. This reaggregation gives rise to self-organised spheres which resemble in vivo oesophageal tissue organisation (106). There are a variety of multicellular spheroid culture models (spinner flask suspension culture, liquid overlay culture, hanging drop culture and microfluidic culture), all with their own advantages and disadvantages, these are detailed in the review by Mehta et al. [2012] (107).
Multicellular spheroid culture have been shown to be effective models in drug permeation (107), cell functionality (108), tumour angiogenesis (109), and tumour-immune cell cross talk (110). In the oesophagus specifically, these models have been used to study ESCC-cell interactions (111), sphere formation in OSC-1 and 2 cell lines (98,112) and comparisons of spheroid formation between cell types (113). The associated limitations are the maintenance of uniform spheres, variability in cell type numbers with each sphere, and a lack of standardised methods for drug screening (114).
Sphere formation assays, another type of spheroid model, follow a similar principle to multicellular spheroid culture, but rather than allowing dissociated cells to aggregate and self-organise, multipotent stem cells from tissues are prevented from aggregating, allowing them to propagate and differentiate into single cell derived spheroids (115). This method has allowed research into the proliferation, self-renewal and multipotency of EADC (116,117) and ESCC (118-120) stem cells, which are known for their influence in their respective cancers.
3D organoids
3D organoids build on the ideas of the previous 3D culture models, aiming to accurately replicate in vivo organ architecture, processes, and parameters on a miniature scale. This is typically done using a single or small number of induced pluripotent or embryonic stem cells, embedded in an extracellular matrix hydrogel to support growth (often Matrigel) (121,122). These multipotent cells are allowed to differentiate, propagate and self-organise into a miniature organ-like scaffold containing a vast array of functional cell layer and types (122,123). The vast array of cell lineages and types give an accurate representation of organ parameters and architecture, unlike that of spheroid culture and OTC. Moreover, 3D organoids are much easier to maintain due to their ability to self-renew. In 3D organoid culture, a patient’s own stem cells can be used, which may have significant implications in the future of personalised medicine (122). Research using oesophageal organoids includes development, differentiation/proliferation and therapeutic approaches to EADC (124,125) and ESCC (126-128), oesophageal signalling pathways in GORD (94,129) and eosinophilic oesophagitis (94).
Absorption modelling—epithelial absorption and mucus function
Similarly, there are limitations to modelling of the intestinal digestive mucosa. Nutrient and drug absorption from the digestive tract involves mucus permeation, brush border enzyme activity and transport across the epithelia via passive diffusion, carrier mediated diffusion, active transport, and pinocytosis (Table 5) (135). Brush border enzymes and specific transport mechanisms are summarised in Tables 2-4. The use of in vitro cell models to simulate intestinal absorption is common, but systems such as the Caco-2 monolayer model do not include a mucus layer (136), other approaches are discussed later.
Table 5
| Class | Mechanism | Function |
|---|---|---|
| Carbohydrate | SGLT1 | Na+ dependent uptake of glucose and galactose from the small intestine. Uptake is mediated by a Na+ concentration gradient maintained by an Na+/K+ ATPase pump which creates a high concentration of Na+ ions in the small intestinal lumen, this allows glucose and galactose symport across the membrane. This is an example of secondary active transport (130) |
| GLUT5 | Facilitated diffusion of fructose from a high concentration on the small intestine to a low concentration in the enterocytes (130) | |
| GLUT2 | Basolateral facilitated diffusion of glucose, galactose, and fructose into serum (131) | |
| Peptides | PEPT1 | H+ dependent symport of di and tripeptides into enterocytes from the small intestinal lumen, peptides are then hydrolysed to amino acids in the cells by internal peptidases (130) |
| Amino acids | ASC | Na+ dependent antiport uptake of alanine, serine, cystine, threonine, glutamine and asparagine (132) |
| B0 | Na+ dependent symport uptake of neutral L-amino acids (130,132) | |
| B0,+ | Na+ and Cl- dependent symport of neutral and cationic amino acids as well as β-ala (130,132) | |
| b0,+ | Independent uptake of cysteine, neutral amino acids and cationic amino acids (130) | |
| β | Apical and basolateral Na+ and Cl-dependent transport (132) of taurine and β-alanine (130,132) | |
| Gly | Na+ and Cl-dependent basolateral symport of glycine into serum from enterocytes (132) | |
| IMINO | Na+ and Cl-dependent symport uptake of proline, hydroxyproline (132) and pipercholic acid (130) | |
| L | Basolateral transport of neutral amino acids (except proline) from enterocytes into serum (132) | |
| N | Na+ (symport) and H+ (antiport) (132) dependent uptake of glutamine, asparagine and histidine (130) bro | |
| PAT | H+ dependent symport uptake of proline, glycine and alanine (130,132) | |
| T | Facilitated diffusion uptake of aromatic amino acids phenylalanine, tyrosine and tryptophan (132) | |
| X-AG | Na+, H+ (symport) and K+(antiport) dependent uptake of (anionic) aspartate and glutamate (132) | |
| Y+L | Na+ dependent cotransport uptake with neutral amino acids (132) | |
| Lipids | CD36 | Facilitates fatty acid uptake from the small intestinal lumen (133) |
| NPC1L1 | Cholesterol uptake transporter (130) | |
| ABCG5, ABCG8 | Efflux of cholesterol from enterocytes into small intestinal lumen (130) | |
| Vitamins | SVCT1, SVCT2 | Na+ dependent vitamin C uptake via Na+ cotransport (130) |
| SMVT | Na+ dependent uptake of biotin by enterocytes (130) | |
| Cubam | Not a transporter as such, but a receptor composed of amnionless and cubulin, responsible for binding the intrinsic factor/vitamin b12 complex, allowing endosome uptake (134) | |
| FOLT, PCFT/HCP1, FOLR1 | Small intestinal folate uptake occurs through these three transporter proteins (130) | |
| RFVT1, RFVTR | Small intestinal uptake of riboflavin is thought to occur using these two transporters (130) | |
| THTR1, THTR2 | Small intestinal thiamine uptake occurs using these two transporters (130) | |
| Other vitamins | Fat soluble vitamins, A, D, K and E typically follow the same route as lipids, which are organised into micelles and absorbed (130) |
Mucus layers coat the epithelia and protect from microbial and enzymatic damage, while also allowing absorption of nutrients. The presence of a mucus layer does however, limit the absorption of certain nanoparticles and drugs in vivo (88). With mucus being a limiting factor for absorption in vivo, it is essential that the mucus layer is accounted for in absorption models.
Mucus is a negatively charged hydrogel, and is typically ~95% water with the glycoprotein mucin (Table 6) being the gel forming component (88,139,140). Mucins are categorised into either 2 subgroups, membrane tethered and secreted (139,141-143). Mucins are high molecular weight glycoproteins varying in size, domain organisation and function—despite this, there are common structural features. The protein backbone of the mucin glycoproteins consists of serine, threonine and proline (STP) repeats. These STP repeats are the fingerprint of mucin molecules. Each mucin type has a unique STP repeat sequence known as a variable number tandem repeat (VNTR) region (88,139,142,144). These are important regions which function as the site of carbohydrate chain attachment, primarily by O-glycosylation, which make up ~70% of the mucin molecular weight (145). The presence of carbohydrate chains on secreted mucins gives them the ability to form gels (146). O-glycosylation occurs when N-acetyl galactosamine (GalNAc) residues are covalently linked to serine or threonine by glucotransferases via an OH group (147). A progressive elongation of the carbohydrate chain, by Golgi glucosyltransferases, up to 20 residues in length occurs using four other carbohydrate residues: N-acetyl glucosamine, fucose, galactose and sialic acid (88,139,144). Proline’s role in the VNTR region is thought to be assistance in the linkage of GalNac residues to serine and threonine (147). Statistical analysis has noted a much higher number of proline residues surrounding glycosylated serine and threonine residues when compared to their non-glycosylated counterparts (148). with affinities depending on proline’s proximity to the O-glycosylation site, with proline at +3 and –1 strongly favouring O-glycosylation as opposed to proline at other sites (149).
Table 6
| Name | Function |
|---|---|
| Mucin | Mucins are heavily glycosylated glycoproteins which are the major component of secreted mucus, a heterogeneous hydrogel which protects the epithelial layers of the gastrointestinal tract from chemical, mechanical, and pathogenic damage, while also providing an essential nutrient source for intestinal flora. Mucins also come in membrane tethered forms which have roles in epithelial protection, the immune response and cell signalling (88) |
| Intrinsic Factor | Secreted by gastric parietal cells, intrinsic factor is a glycoprotein which is essential for the absorption of cobalamin (vitamin B12). Intrinsic factor is present in the small intestine when cobalamin is release from its protein complex and becomes available for binding. The cobalamin/intrinsic factor complex binds to receptors on the epithelial cells of the terminal ileum, prompting absorption (137) |
| Bile | Bile, conjugated in the liver (as bile acids) and stored in the gall bladder, are an essential component of lipid absorption. Lipids are not water soluble; this makes the surface area for enzymatic action low. Bile works by emulsifying lipids, creating an oil: water interface at which pancreatic lipase and its cofactor, colipase, allows effective digestions of lipids into their simple forms, where they can be absorbed (138) |
Carbohydrate side chains are heavily sulphated upon elongation—it is their presence, alongside sialic acid, which allow mucus to have a negative charge at physiological pH levels (88).
Secreted mucins (Figure 3; Table 7), provide a mucus layer for several mucosal surfaces. Of these, MUC2, was one of the first secreted mucins to be characterised in detail (88,139,151-158). MUC2 (Figure 3) is the predominant secreted mucin of the SI (88,152,159,160) and the colon (152,157,158). At the N-terminal, vWF-like domains D1, D2, D’ and D3 are found, while at the C-terminal a vWF-like D4 domain is found, followed by a B, C and CK domain. The central region of MUC2 is split into two VNTR regions, a short region closer to the N-terminal which is flanked by 2 globular cysteine domains (CYS) domains at either side, and a longer region which spans most of the backbone. These regions are separated by the single globular CYS domain found at the C-terminal side of the first VNTR region (88,151,157). The initial short VNTR region is conserved and shows little variability in the number of STP repeats which are generally 23 amino acids long (151). The longer VNTR region however is highly variable with 40–185 STP repeats (151,161) which contributes to its large size of up to 7 mDa (155).
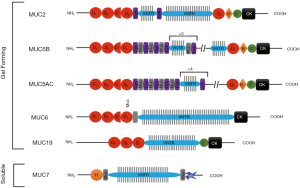
Table 7
| Mucin gene | Secreted/membrane | Tissue expression | Chromosomal location |
|---|---|---|---|
| MUC 1 | Membrane | All epithelia, breast, pancreas, small intestine, bladder | 1q21 |
| MUC 2 | Secreted (gel-forming) | Colon, small intestine, airways | 11p15.5 |
| MUC 3B | Membrane | Colon, small intestine, bladder | 7q22 |
| MUC 3A | Membrane | Colon, small intestine, bladder heart, liver, thymus, pancreas | 7q22 |
| MUC 4 | Membrane | Airways, colon, small intestine, stomach, cervix, eye | 3q29 |
| MUC 5AC | Secreted (gel-forming) | Airways, stomach, cervix, middle ear, eye | 11p15.5 |
| MUC 5B | Secreted (gel-forming) | Airways, submaxillary gland, cervix, gallbladder, middle ear | 11p15.5 |
| MUC 6 | Secreted (gel-forming) | Stomach, gall bladder, cervix | 11p15.5 |
| MUC 7 | Secreted (soluble) | Salivary glands, airways, eye | 4q13-21 |
| MUC 8 | Membrane | Airways | 12q24.3 |
| MUC 9 | Membrane | Oviduct | 1p13 |
| MUC 12 | Membrane | Colon | 7q22 |
| MUC 13 | Membrane | GI tract, colon, airways | 3q13.3 |
| MUC 15 | Membrane | Colon, airways, small intestine, spleen, prostate, breast, eye | 11p14.3 |
| MUC 16 | Membrane | Ovarian epithelial cells, nasal mucosa, eye | 19p13.3 |
| MUC 17 | Membrane | Duodenum, stomach, colon | 7q22 |
| MUC 18 | Membrane | Lung, breast | 11q23 |
| MUC 19 | Secreted (gel-forming) | Salivary glands, trachea submucosal glands | 12q12 |
| MUC 20 | Membrane | Kidney placenta, colon, lung, eye, prostate, liver, eye | 3q29 |
| MUC 21 | Membrane | Lung, colon, thymus | 6p21.32 |
| MUC 22 | Membrane | Lung | 6p21.31 |
The mechanism of secretion of MUC2 has been well characterised and is described in detail by Pearson et al. [2016] (88), Ambort et al. [2011; 2012] (153,154) and Johansson et al. [2011] (157). In the endoplasmic reticulum at pH 7.2, the MUC2 monomers are linked in trios at the D3 domain via disulphide bridges, creating MUC2 trimers. These trimers are transported to the Golgi Apparatus where the bulk of O-glycosylation takes place. In the Golgi Apparatus, the pH drops to 5.2—this causes protonation of histidine residues which in turn allows non covalent interactions between the vWF-like domains D1, D2 and D3, creating 5 or 6 sided ring-like structures which operate with D3 as the corners. An increase in Ca2+ levels in the Golgi Apparatus allows cross linking of negative charges, allowing the mucins to be tightly packed into secretory granules. A final dimerisation at the CK domain ensures that the mucin polymers are ready for secretion. Upon secretion, Na+ ions displace Ca2+ ions, uncoupling the cross links—this allows mucus to rapidly hydrate and swell 1,000–3,000-fold.
MUC2 polymers are arranged into 5- or 6-sided ring like structures which act as pores. Research suggests these pores vary in size due to the sliding of the mucus layers but are understood to have a size limit of roughly 200 nm in diameter (88). The CYS domains are thought to determine their pore size through non-covalent bonds between half-cysteine residues and adjacent, positively charged amino acids (154). These pores allow particles smaller than 200 nm to traverse the mucus layer and size exclude larger particles. With no mucus layer, particles larger than 200 nm will easily access the epithelia. Moreover, the strong negative charges will cause mucus/particle interactions. Small neutral particles will likely cross the mucus layer with little hindrance. Negatively charged particles will likely be repelled by the mucus layers negatively charged residues, and positively charged particles will bind negatively charged residues and become trapped.
Approaches to mucus modelling
Groo et al. [2014] identified that although many methods for mucus diffusion studies are available there is not a standard consensus protocol (162). Several in vitro and ex vivo approaches have been adopted to mucus modelling, some of which are integrated with cell-culture systems, and some which are non-integrated and model the mucus layer independently.
Li et al. [2013] described the lack of mucus modelling as a limitation of Caco-2 monoculture systems, and although they do express membrane bound MUC1, they do not produce secreted intestinal mucins (136). As an advance on CACO-2 monoculture, co-culture of Caco-2 and mucus producing HT29 cells have been developed. HT29-MTX cells only secrete gastric type MUC5AC, whereas HT29-FU cells adapted with fluorouracil secrete intestinal type mucin MUC2 (163). While a key advantage of co-culture systems is that the mucin is directly secreted above, and in intimate contact with the epithelia, the secreted mucin layer is not complete, and with a reported thickness of ~4 µm, which does not recapitulate an in vivo human mucus layer (which is two orders of magnitude thicker) (164). To allow for full integration of epithelial culture systems with digestive models, it is essential that the mucus layer retains relevant permeation characteristics, and can protect the underlying epithelia from components of digestive secretions. To our knowledge there is no published data on Caco-2-HT29-compatibility with digestive secretions, however it would be surprising if the thin mucin covering produced in co-culture would be sufficient to achieve either of these requirements. Despite limitations, the co-culture model is reported to generate more predictable experimental results compared to monoculture, and co-culture systems have been advanced further to integrate intestinal M-Like cells in a triple culture system (165).
Mucus/mucin source is one of the key considerations, and Lock et al. discuss the importance of factors including species, age, state of health, anatomical site of collection and inter-donor (animal or human) variability (163). Inter-donor variation can be somewhat overcome by pooling mucus from multiple donors prior to testing, and that samples can be stored frozen without significant changes to mucus rheology.
We have previously argued that while many studies have used mucins to simulate mucus function, it is important that these mucins replicate the native state (88). Sigma purified mucin is one of the most widely available commercial sources of mucin, but due to proteolytic degradation during purification is not structurally comparable to native mucin, cannot form a gel, and affects permeation characteristics (88,163).
Attempts have been made to develop ‘biosimilar’ mucus using polyacrylic acid (carbopol), BSA and linoleic acid in combination with Sigma porcine mucin to make the viscoelastic properties and microstructure more comparable to native mucus. While lipid and protein components are normal components of the mucus gel, polyacrylic acid is not, and significant research has been conducted to show mucin-carbopol interactions can bring about dramatic changes to viscosity (166). Because of its mucus modifying properties and inhibition of pepsin hydrolysis of mucus, carbopol has been investigated as a muco-protective agent (88).
In order to better replicate mucus thickness, our group in collaboration, developed a transmembrane permeation system using native porcine small intestinal mucus in a transwell set up with a resulting mucus layer of a thickness of 929±115 µm (167). While this is thicker than data reported for human small intestinal mucus thickness, the thickness is within the same order of magnitude, and allowed for a higher throughput, reproducible method from which time-course data could be collected, and permeation data can be normalised to in vivo mucus thickness. This has advantages over previous destructive methods which involve permeating compounds through a mucus filled device, and then taking sections in order to track compound permeation at a single timepoint.
Other approaches to mucus permeation include side-on three-compartment diffusion chambers, diffusion cells and Ussing chambers (162). A limitation of these systems can be mucus thickness, for example Bhat et al. used a 3,000 µm thick chamber which is considerably higher than in vivo, whereas Norris and Sinko used a mucus thickness of 380 µm. Furthermore Bhat et al. showed that the membrane material can cause significant variability in permeability data (168,169). This is a further advantage of using integrated systems that bring the mucus layer into direct contact with digestive secretions, and model epithelia.
While ex vivo studies of tissue samples can be limited by availability of material and are subject to donor variation, they naturally have advantages in terms of modelling the tissue architecture, and mucus thickness and composition (163). Intestinal loops and Ussing chambers are common methods for ex vivo tissue modelling.
Discussions and future directions
All 3D models described in this paper have proven to be useful in oesophageal research, with the 3D organoid presenting the most promising model, with a huge potential in personalised medicine. However, there is room for improvement, and this should be considered when further developing these models. While all models described may not contain all cells representative of a specific organ, and cell representation is somewhat variable between each culture, there is another major limiting factor - the lack of other, functional layers found deeper within the oesophageal tissue structure. This paper has already discussed the importance of the smooth muscle in the functions of the oesophagus, but there are important functions for immune cells, stromal cell types, neuronal control and a vascular system for nutrient transport and waste removal (92,114) which are not considered. Moreover, these deeper tissue layers can be subject to disease, such as ESCC or EADC tumour invasion (92). It is important to develop more complex oesophageal models in the future, which consider the latter points and contribute to a better mimicking of human oesophageal function, both in healthy and diseased oesophagi. There have been some successful developments in this area, with an example coming from Workman et al. [2017], who developed a human pluripotent-stem-cell-derived intestinal model to include a functional enteric nervous system by combining neural crest cells, which initiate mesenchymal differentiation into neuronal structures, and human intestinal organoids (129). Despite this model focusing on modelling the intestine, it shows promise for the utilisation of similar methods in the development of more complex oesophageal models.
An important area of future research will be to evaluate current and emerging models for use in modelling the action of reflux and the role this plays in disease pathology.
Through this paper we have identified limitations of modelling of the oesophageal and intestinal mucosa. Current research in our lab is based around development of fully integrated models that recapitulate the protective function and permeation characteristics of mucus to allow modelling of whole digestive secretions in cell culture systems that integrate a cell-compatible mucus barrier.
Acknowledgments
Funding: This work was supported by the BBSRC Super Follow on Fund (BB/R019657/1).
Footnote
Provenance and Peer Review: This article was commissioned by the editorial office, Annals of Esophagus for the series “Epidemiology, Biomarkers and Modelling of Gastroesophageal Reflux Disease”. The article has undergone external peer review.
Reporting Checklist: The authors have completed the Narrative Review reporting checklist. Available at https://aoe.amegroups.com/article/view/10.21037/aoe-20-90/rc
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://aoe.amegroups.com/article/view/10.21037/aoe-20-90/coif). The series “Epidemiology, Biomarkers and Modelling of Gastroesophageal Reflux Disease” was commissioned by the editorial office without any funding or sponsorship. JP served as the unpaid Guest Editor of the series. PC reports a financial interest in Aelius Biotech, and has a patent EP3074769A2 issued. MW reports other from Aelius Biotech, outside the submitted work, and has a patent WO2015075467A3 issued. JP reports other from Aelius Biotech, outside the submitted work, and has a patent WO2015075467A3 issued, and a patent EP3074769A2 issued. The authors have no other conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Sweis R. Physiology and function of the oesophagus. In: Lomer M, editor. Advanced Nutrition and Dietetics in Gastroenterology. John Wiley & Sons, Ltd.; 2014.
- Aziz Q, Fass R, Gyawali CP, et al. Functional Esophageal Disorders. Gastroenterology 2016; [Epub ahead of print]. [Crossref] [PubMed]
- Alegria A, Garcia-Llatas G, Cilla A. Static Digestion Models: General Introduction. In: Verhoeckx K, Cotter P, Lopez-Exposito I, et al. editors. The Impact of Food Bioactives on Health: in vitro and ex vivo models. Cham (CH); 2015:3-12.
- Thuenemann EC. Dynamic Digestion Models: General Introduction. In: Verhoeckx K, Cotter P, Lopez-Exposito I, et al. editors. The Impact of Food Bioactives on Health: in vitro and ex vivo models. Cham (CH); 2015:33-6.
- Lea T. Caco-2 cell line. The impact of food bioactives on health. Springer, Cham; 2015:103-11.
- Lea T. Epithelial Cell Models; General Introduction. In: Verhoeckx K, Cotter P, Lopez-Exposito I, et al. editors. The Impact of Food Bioactives on Health: in vitro and ex vivo models. Cham (CH); 2015:95-102.
- Costa J, Ahluwalia A. Advances and Current Challenges in Intestinal in vitro Model Engineering: A Digest. Front Bioeng Biotechnol 2019;7:144. [Crossref] [PubMed]
- Ripken D, Hendriks HFJ. Porcine Ex Vivo Intestinal Segment Model. In: Verhoeckx K, Cotter P, Lopez-Exposito I, et al. editors. The Impact of Food Bioactives on Health: in vitro and ex vivo models. Cham (CH); 2015:255-62.
- McDonald JAK. In vitro models of the human microbiota and microbiome. Emerg Top Life Sci 2017;1:373-84. [Crossref] [PubMed]
- Mattei G GS, Ahluwalia A. Design criteria for generating physiologically relevant in vitro models in bioreactors. Processes 2014;2:548-69. [Crossref]
- Dupont D, Alric M, Blanquet-Diot S, et al. Can dynamic in vitro digestion systems mimic the physiological reality? Crit Rev Food Sci Nutr 2019;59:1546-62. [Crossref] [PubMed]
- Lefebvre DE, Venema K, Gombau L, et al. Utility of models of the gastrointestinal tract for assessment of the digestion and absorption of engineered nanomaterials released from food matrices. Nanotoxicology 2015;9:523-42. [Crossref] [PubMed]
- Lin L, Wong H. Predicting Oral Drug Absorption: Mini Review on Physiologically-Based Pharmacokinetic Models. Pharmaceutics 2017;9:41. [Crossref] [PubMed]
- Astashkina A, Mann B, Grainger DW. A critical evaluation of in vitro cell culture models for high-throughput drug screening and toxicity. Pharmacol Ther 2012;134:82-106. [Crossref] [PubMed]
- Cordonnier C, Thévenot J, Etienne-Mesmin L, et al. Dynamic In Vitro Models of the Human Gastrointestinal Tract as Relevant Tools to Assess the Survival of Probiotic Strains and Their Interactions with Gut Microbiota. Microorganisms 2015;3:725-45. [Crossref] [PubMed]
- Dupont D, Mackie AR. Static and dynamic in vitro digestion models to study protein stability in the gastrointestinal tract. Drug Discovery Today: Disease Models 2015;1:23-7. [Crossref]
- Shoji Y, Nakashima H. Nutraceutics and delivery systems. J Drug Target 2004;12:385-91. [Crossref] [PubMed]
- Minekus M, Alminger M, Alvito P, et al. A standardised static in vitro digestion method suitable for food - an international consensus. Food Funct 2014;5:1113-24. [Crossref] [PubMed]
- Kulkarni BV, Mattes RD. Lingual lipase activity in the orosensory detection of fat by humans. Am J Physiol Regul Integr Comp Physiol 2014;306:R879-85. [Crossref] [PubMed]
- Mishra PJ, Ragunath C, Ramasubbu N. The mechanism of salivary amylase hydrolysis: Role of residues at subsite S2′. Biochem Biophys Res Commun 2002;292:468-73. [Crossref] [PubMed]
- Valls C, Rojas C, Pujadas G, et al. Characterization of the activity and stability of amylase from saliva and detergent: laboratory practicals for studying the activity and stability of amylase from saliva and various commercial detergents. Biochem Mol Biol Educ 2012;40:254-65. [Crossref] [PubMed]
- Bardhan KD, Strugala V, Dettmar PW. Reflux revisited: advancing the role of pepsin. Int J Otolaryngol 2012;2012:646901. [Crossref] [PubMed]
- Fruton JS. The mechanism of the catalytic action of pepsin and related acid proteinases. Adv Enzymol Relat Areas Mol Biol 1976;44:1-36. [Crossref] [PubMed]
- Hamosh M. Lingual and gastric lipases. Nutrition 1990;6:421-8. [PubMed]
- Abrams CK, Hamosh M, Lee TC, et al. Gastric lipase: localization in the human stomach. Gastroenterology 1988;95:1460-4. [Crossref] [PubMed]
- Trypsin RW. Methods of enzymatic analysis. Elsevier; 1974:1013-24.
- Appel W. Chymotrypsin: molecular and catalytic properties. Clin Biochem 1986;19:317-22. [Crossref] [PubMed]
- Appel W. Carboxypeptidases. Methods of enzymatic analysis. Elsevier; 1974:986-8.
- Greene D, Das B, Fricker LD. Regulation of carboxypeptidase E. Effect of pH, temperature and Co2+ on kinetic parameters of substrate hydrolysis. Biochem J 1992;285:613-8. [Crossref] [PubMed]
- de Oliveira EB, Salgado MCO. Pancreatic elastases. Handbook of Proteolytic Enzymes. Elsevier; 2013:2639-45.
- Starkey PM, Barrett AJ. Human lysosomal elastase. Catalytic and immunological properties. Biochem J 1976;155:265-71. [Crossref] [PubMed]
- Lowe ME. Structure and function of pancreatic lipase and colipase. Annu Rev Nutr 1997;17:141-58. [Crossref] [PubMed]
- Nevalainen TJ. The role of phospholipase A in acute pancreatitis. Scand J Gastroenterol 1980;15:641-50. [Crossref] [PubMed]
- Grataroli R, Dijkman R, Dutilh CE, et al. Studies on Prophospholipase A2 and Its Enzyme from Human Pancreatic Juice: Catalytic Properties and Sequence of the N‐Terminal Region. Eur J Biochem 1982;122:111-7. [Crossref] [PubMed]
- Williams JA. Regulation of acinar cell function in the pancreas. Curr Opin Gastroenterol 2010;26:478-83. [Crossref] [PubMed]
- Sky-Peck HH, Thuvasethakul P. Human pancreatic alpha-amylase. II. Effects of pH, substrate and ions on the activity of the enzyme. Ann Clin Lab Sci 1977;7:310-7. [PubMed]
- Holmes R. Carbohydrate digestion and absorption. J Clin Pathol Suppl (R Coll Pathol) 1971;5:10. [Crossref]
- Feher JJ. Quantitative human physiology: an introduction. Academic Press; 2017:823-4.
- Tobey N, Heizer W, Yeh R, et al. Human intestinal brush border peptidases. Gastroenterology 1985;88:913-26. [Crossref] [PubMed]
- Holmes R, Lobley RW. Intestinal brush border revisited. Gut 1989;30:1667-78. [Crossref] [PubMed]
- Liddle RA. Cholecystokinin cells. Annu Rev Physiol 1997;59:221-42. [Crossref] [PubMed]
- Burtis CA, Ashwood ER, Bruns DE. Tietz textbook of clinical chemistry and molecular diagnostics-e-book. Elsevier Health Sciences; 2012:1719-20.
- Liu Y, Vosmaer GD, Tytgat GN, et al. Gastrin (G) cells and somatostatin (D) cells in patients with dyspeptic symptoms: Helicobacter pylori associated and non-associated gastritis. J Clin Pathol 2005;58:927-31. [Crossref] [PubMed]
- Afroze S, Meng F, Jensen K, et al. The physiological roles of secretin and its receptor. Ann Transl Med 2013;1:29. [PubMed]
- Iwasaki M, Akiba Y, Kaunitz JD. Recent advances in vasoactive intestinal peptide physiology and pathophysiology: focus on the gastrointestinal system. F1000Res 2019;8:eF1000 Faculty Rev-1629.
- McIntosh CH, Widenmaier S, Kim SJ. Glucose‐dependent insulinotropic polypeptide (gastric inhibitory polypeptide; GIP). Vitam Horm 2009;80:409-71. [Crossref] [PubMed]
- Pedersen AM, Bardow A, Jensen SB, et al. Saliva and gastrointestinal functions of taste, mastication, swallowing and digestion. Oral Dis 2002;8:117-29. [Crossref] [PubMed]
- Thuenemann EC, Mandalari G, Rich GT, et al. Dynamic gastric model (DGM). The impact of food bioactives on health. Springer, Cham; 2015:47-59.
- Kong F, Singh RP. A human gastric simulator (HGS) to study food digestion in human stomach. J Food Sci 2010;75:E627-35. [Crossref] [PubMed]
- Kozu H, Nakata Y, Nakajima M, et al. Development of a human gastric digestion simulator equipped with peristalsis function for the direct observation and analysis of the food digestion process. Food Science and Technology Research 2014;20:225-33. [Crossref]
- Marciani L, Gowland PA, Spiller RC, et al. Effect of meal viscosity and nutrients on satiety, intragastric dilution, and emptying assessed by MRI. Am J Physiol Gastrointest Liver Physiol 2001;280:G1227-33. [Crossref] [PubMed]
- Vardakou M, Mercuri A, Barker SA, et al. Achieving antral grinding forces in biorelevant in vitro models: comparing the USP dissolution apparatus II and the dynamic gastric model with human in vivo data. AAPS PharmSciTech 2011;12:620-6. [Crossref] [PubMed]
- Marciani L, Gowland PA, Fillery-Travis A, et al. Assessment of antral grinding of a model solid meal with echo-planar imaging. Am J Physiol Gastrointest Liver Physiol 2001;280:G844-9. [Crossref] [PubMed]
- Venema K. The TNO in vitro model of the colon (TIM-2). The Impact of Food Bioactives on Health. Springer, Cham; 2015:293-304.
- Minekus M, Verhoeckx KCotter P, et al. The TNO Gastro-Intestinal Model (TIM) 2015.
- Minekus M, Havenaar R. In vitro model of an in vivo digestive tract. Google Patents; 1996.
- Minekus M, Marteau P, Havenaar R. Multicompartmental dynamic computer-controlled model simulating the stomach and small intestine. Alternatives to laboratory animals: ATLA 1995.
- Ménard O, Cattenoz T, Guillemin H, et al. Validation of a new in vitro dynamic system to simulate infant digestion. Food Chem 2014;145:1039-45. [Crossref] [PubMed]
- Guerra A, Denis S, le Goff O, et al. Development and validation of a new dynamic computer‐controlled model of the human stomach and small intestine. Biotechnol Bioeng 2016;113:1325-35. [Crossref] [PubMed]
- Lamb PJ, Griffin SM. The anatomy and physiology of the oesophagus. Upper Gastrointestinal Surgery. Springer; 2005:1-15.
- Mashimo H, Goyal RK. Physiology of esophageal motility. GI Motility online; 2006.
- Chaudhry SR, Bordoni B. Anatomy, Thorax, Esophagus. StatPearls [Internet]. StatPearls Publishing; 2019.
- Kuo B, Urma D. Esophagus-anatomy and development. GI Motility online; 2006.
- Orlando RC, Lacy ER, Tobey NA, et al. Barriers to paracellular permeability in rabbit esophageal epithelium. Gastroenterology 1992;102:910-23. [Crossref] [PubMed]
- Orlando RC. Esophageal mucosal defense mechanisms. GI Motility online; 2006.
- Orlando RC. The integrity of the esophageal mucosa. Balance between offensive and defensive mechanisms. Best Pract Res Clin Gastroenterol 2010;24:873-82. [Crossref] [PubMed]
- Günther C, Neumann H, Vieth M. Esophageal epithelial resistance. Dig Dis 2014;32:6-10. [Crossref] [PubMed]
- Tracht J, Robinson BS, Krasinskas AM. Pathology of Premalignant and Malignant Disease of the Esophagus. Esophageal Cancer. Springer; 2020:61-81.
- Wang DH, Souza RF. Biology of Barrett's esophagus and esophageal adenocarcinoma. Gastrointest Endosc Clin N Am 2011;21:25-38. [Crossref] [PubMed]
-
Mittal RK 2011 . - Long JD, Orlando RC. Esophageal submucosal glands: structure and function. Am J Gastroenterol 1999;94:2818-24. [Crossref] [PubMed]
- Guillem P, Billeret V, Buisine MP, et al. Mucin gene expression and cell differentiation in human normal, premalignant and malignant esophagus. Int J Cancer 2000;88:856-61. [Crossref] [PubMed]
- Arul GS, Moorghen M, Myerscough N, et al. Mucin gene expression in Barrett's oesophagus: an in situ hybridisation and immunohistochemical study. Gut 2000;47:753-61. [Crossref] [PubMed]
- Davies JR, Svitacheva N, Lannefors L, et al. Identification of MUC5B, MUC5AC and small amounts of MUC2 mucins in cystic fibrosis airway secretions. Biochem J 1999;344:321-30. [Crossref] [PubMed]
- Kesimer M, Makhov AM, Griffith JD, et al. Unpacking a gel-forming mucin: a view of MUC5B organization after granular release. Am J Physiol Lung Cell Mol Physiol 2010;298:L15-22. [Crossref] [PubMed]
- Kirkham S, Sheehan JK, Knight D, et al. Heterogeneity of airways mucus: variations in the amounts and glycoforms of the major oligomeric mucins MUC5AC and MUC5B. Biochem J 2002;361:537-46. [Crossref] [PubMed]
- Dixon J, Strugala V, Griffin S, et al. Esophageal mucin: an adherent mucus gel barrier is absent in the normal esophagus but present in columnar-lined Barrett’s esophagus. Am J Gastroenterol 2001;96:2575-83. [Crossref] [PubMed]
- Sivarao DV, Goyal RK. Functional anatomy and physiology of the upper esophageal sphincter. Am J Med 2000;108:27S-37S. [Crossref] [PubMed]
- Mittal RK, Balaban DH. The esophagogastric junction. N Engl J Med 1997;336:924-32. [Crossref] [PubMed]
- Farré R, Sifrim D. Regulation of basal tone, relaxation and contraction of the lower oesophageal sphincter. Relevance to drug discovery for oesophageal disorders. Br J Pharmacol 2008;153:858-69. [Crossref] [PubMed]
- Mittal RK, Goyal RK. Sphincter mechanisms at the lower end of the esophagus. GI Motility online; 2006.
- Richter JE. Achalasia and lower esophageal sphincter anatomy and physiology: Implications for peroral esophageal myotomy technique. Tech Gastrointest Endosc 2013;15:122-6. [Crossref]
- Higgs B, Shorter RG, Ellis FH. A study of the anatomy of the human esophagus with special reference to the gastroesophageal sphincter. J Surg Res 1965;5:503-7. [Crossref]
- Sidhu AS, Triadafilopoulos G. Neuro-regulation of lower esophageal sphincter function as treatment for gastroesophageal reflux disease. World J Gastroenterol 2008;14:985-90. [Crossref] [PubMed]
- Liebermann-Meffert D, Allgöwer M, Schmid P, et al. Muscular equivalent of the lower esophageal sphincter. Gastroenterology 1979;76:31-8. [Crossref] [PubMed]
- Forssell H. Gastric mucosal defence mechanisms: a brief review. Scand J Gastroenterol Suppl 1988;155:23-8. [Crossref] [PubMed]
- Quigley E, Turnberg L. pH of the microclimate lining human gastric and duodenal mucosa in vivo: studies in control subjects and in duodenal ulcer patients. Gastroenterology 1987;92:1876-84. [Crossref] [PubMed]
- Pearson JP, Chater PI, Wilcox MD. The properties of the mucus barrier, a unique gel–how can nanoparticles cross it? Ther Deliv 2016;7:229-44. [Crossref] [PubMed]
- Pelaseyed T, Zäch M, Petersson ÅC, et al. Unfolding dynamics of the mucin SEA domain probed by force spectroscopy suggest that it acts as a cell‐protective device. FEBS J 2013;280:1491-501. [Crossref] [PubMed]
- Lindén SK, Sheng YH, Every AL, et al. MUC1 limits Helicobacter pylori infection both by steric hindrance and by acting as a releasable decoy. PLoS Pathog 2009;5:e1000617. [Crossref] [PubMed]
- Tétreault MP. Esophageal Cancer: Insights From Mouse Models. Cancer Growth Metastasis 2015;8:37-46. [PubMed]
- Shacham-Silverberg V, Wells JM. Generation of esophageal organoids and organotypic raft cultures from human pluripotent stem cells. Methods Cell Biol 2020;159:1-22. [Crossref] [PubMed]
- Spechler SJ. Of Mice and Men and Metaplasia. Cell Mol Gastroenterol Hepatol 2017;4:183-4. [Crossref] [PubMed]
- Kasagi Y, Chandramouleeswaran PM, Whelan KA, et al. The Esophageal Organoid System Reveals Functional Interplay Between Notch and Cytokines in Reactive Epithelial Changes. Cell Mol Gastroenterol Hepatol 2018;5:333-52. [Crossref] [PubMed]
- Nunes AS, Barros AS, Costa EC, et al. 3D tumor spheroids as in vitro models to mimic in vivo human solid tumors resistance to therapeutic drugs. Biotechnol Bioeng 2019;116:206-26. [Crossref] [PubMed]
- Kalabis J, Wong GS, Vega ME, et al. Isolation and characterization of mouse and human esophageal epithelial cells in 3D organotypic culture. Nat Protoc 2012;7:235-46. [Crossref] [PubMed]
- Shamir ER, Ewald AJ. Three-dimensional organotypic culture: experimental models of mammalian biology and disease. Nat Rev Mol Cell Biol 2014;15:647-64. [Crossref] [PubMed]
- Andl CD, Fargnoli BB, Okawa T, et al. Coordinated functions of E-cadherin and transforming growth factor β receptor II in vitro and in vivo. Cancer Res 2006;66:9878-85. [Crossref] [PubMed]
- Trisno SL, Philo KED, McCracken KW, et al. Esophageal Organoids from Human Pluripotent Stem Cells Delineate Sox2 Functions during Esophageal Specification. Cell Stem Cell 2018;23:501-515.e7. [Crossref] [PubMed]
- Karakasheva TA, Lin EW, Tang Q, et al. IL-6 Mediates Cross-Talk between Tumor Cells and Activated Fibroblasts in the Tumor Microenvironment. Cancer Res 2018;78:4957-70. [Crossref] [PubMed]
- Loomans HA, Arnold SA, Quast LL, et al. Esophageal squamous cell carcinoma invasion is inhibited by Activin A in ACVRIB-positive cells. BMC Cancer 2016;16:873. [Crossref] [PubMed]
- Yang X, Lehman H, Bruggeman R, et al. Esophageal squamous cell carcinoma model of 3D organotypic tissue culture system with genetic modification of epithelial growth factor receptor and p120‐catenin (1119.3). FASEB J 2014;28:1119.3.
- Kong J, Crissey MA, Stairs DB, et al. Cox2 and β-catenin/T-cell factor signaling intestinalize human esophageal keratinocytes when cultured under organotypic conditions. Neoplasia 2011;13:792-805. [Crossref] [PubMed]
- Zhou Z, Lu H, Zhu S, et al. Activation of EGFR-DNA-PKcs pathway by IGFBP2 protects esophageal adenocarcinoma cells from acidic bile salts-induced DNA damage. J Exp Clin Cancer Res 2019;38:13. [Crossref] [PubMed]
- Underwood TJ, Derouet MF, White MJ, et al. A comparison of primary oesophageal squamous epithelial cells with HET‐1A in organotypic culture. Biology of the Cell 2010;102:635-44. [Crossref] [PubMed]
- Katt ME, Placone AL, Wong AD, et al. In Vitro Tumor Models: Advantages, Disadvantages, Variables, and Selecting the Right Platform. Front Bioeng Biotechnol 2016;4:12. [Crossref] [PubMed]
- Mehta G, Hsiao AY, Ingram M, et al. Opportunities and challenges for use of tumor spheroids as models to test drug delivery and efficacy. J Control Release 2012;164:192-204. [Crossref] [PubMed]
- Hirschhaeuser F, Menne H, Dittfeld C, et al. Multicellular tumor spheroids: an underestimated tool is catching up again. J Biotechnol 2010;148:3-15. [Crossref] [PubMed]
- Mueller-Klieser W. Method for the determination of oxygen consumption rates and diffusion coefficients in multicellular spheroids. Biophys J 1984;46:343-8. [Crossref] [PubMed]
- Gottfried E, Kunz-Schughart LA, Andreesen R, et al. Brave little world: spheroids as an in vitro model to study tumor-immune-cell interactions. Cell Cycle 2006;5:691-5. [Crossref] [PubMed]
- Shima I, Kubota S, Sasaguri Y, et al. Rearrangement of esophageal-carcinoma cells and stromal fibroblasts in a multicellular spheroid. Int J Oncol 1995;7:795-800. [Crossref] [PubMed]
- Sarbia M, Bösing N, Hildebrandt B, et al. Characterization of two newly established cell lines derived from squamous cell carcinomas of the oesophagus. Anticancer Res 1997;17:2185-92. [PubMed]
- Korff T, Augustin HG. Integration of endothelial cells in multicellular spheroids prevents apoptosis and induces differentiation. J Cell Biol 1998;143:1341-52. [Crossref] [PubMed]
- Fang Y, Eglen RM. Three-Dimensional Cell Cultures in Drug Discovery and Development. SLAS Discov 2017;22:456-72. [Crossref] [PubMed]
- Bahmad HF, Cheaito K, Chalhoub RM, et al. Sphere-Formation Assay: Three-Dimensional in vitro Culturing of Prostate Cancer Stem/Progenitor Sphere-Forming Cells. Front Oncol 2018;8:347. [Crossref] [PubMed]
- Wang Z, Da Silva TG, Jin K, et al. Notch signaling drives stemness and tumorigenicity of esophageal adenocarcinoma. Cancer Res 2014;74:6364-74. [Crossref] [PubMed]
- Jiménez P, Chueca E, Arruebo M, et al. CD24 Expression Is Increased in 5-Fluorouracil-Treated Esophageal Adenocarcinoma Cells. Front Pharmacol 2017;8:321. [Crossref] [PubMed]
- Qin G, Lian J, Yue D, et al. Musashi1, a potential prognostic marker in esophageal squamous cell carcinoma. Oncol Rep 2017;38:1724-32. [Crossref] [PubMed]
- Huang L, Lian J, Chen X, et al. WASH overexpression enhances cancer stem cell properties and correlates with poor prognosis of esophageal carcinoma. Cancer Sci 2017;108:2358-65. [Crossref] [PubMed]
- Liu JQ, Deng M, Xue NN, et al. lncRNA KLF3-AS1 Suppresses Cell Migration and Invasion in ESCC by Impairing miR-185-5p-Targeted KLF3 Inhibition. Mol Ther Nucleic Acids 2020;20:231-41. [Crossref] [PubMed]
- Yin X, Mead BE, Safaee H, et al. Engineering Stem Cell Organoids. Cell Stem Cell 2016;18:25-38. [Crossref] [PubMed]
- Perkhofer L, Frappart PO, Müller M, et al. Importance of organoids for personalized medicine. Per Med 2018;15:461-5. [Crossref] [PubMed]
- Lancaster MA, Knoblich JA. Generation of cerebral organoids from human pluripotent stem cells. Nat Protoc 2014;9:2329-40. [Crossref] [PubMed]
- Li X, Francies HE, Secrier M, et al. Organoid cultures recapitulate esophageal adenocarcinoma heterogeneity providing a model for clonality studies and precision therapeutics. Nat Commun 2018;9:2983. [Crossref] [PubMed]
- Hassan MS, Lenga M, Petrova L, et al. Therapeutic targeting of hypoxia in experimental esophageal adenocarcinoma. AACR; 2019.
- Imai T, Oue N, Sentani K, et al. KIF11 Is Required for Spheroid Formation by Oesophageal and Colorectal Cancer Cells. Anticancer Res 2017;37:47-55. [Crossref] [PubMed]
- Zhao Y, Lu Q, Li C, et al. PRMT1 regulates the tumour-initiating properties of esophageal squamous cell carcinoma through histone H4 arginine methylation coupled with transcriptional activation. Cell Death Dis 2019;10:359. [Crossref] [PubMed]
- Pham QT, Oue N, Sekino Y, et al. TDO2 Overexpression Is Associated with Cancer Stem Cells and Poor Prognosis in Esophageal Squamous Cell Carcinoma. Oncology 2018;95:297-308. [Crossref] [PubMed]
- Workman MJ, Mahe MM, Trisno S, et al. Engineered human pluripotent-stem-cell-derived intestinal tissues with a functional enteric nervous system. Nat Med 2017;23:49-59. [Crossref] [PubMed]
- Kiela PR, Ghishan FK. Physiology of Intestinal Absorption and Secretion. Best Pract Res Clin Gastroenterol 2016;30:145-59. [Crossref] [PubMed]
- Drozdowski LA, Thomson AB. Intestinal sugar transport. World J Gastroenterol 2006;12:1657-70. [Crossref] [PubMed]
- Bröer S, Fairweather SJ. Amino Acid Transport Across the Mammalian Intestine. Compr Physiol 2018;9:343-73. [Crossref] [PubMed]
- Pepino MY, Kuda O, Samovski D, et al. Structure-function of CD36 and importance of fatty acid signal transduction in fat metabolism. Annu Rev Nutr 2014;34:281-303. [Crossref] [PubMed]
- Jensen LL, Andersen RK, Hager H, et al. Lack of megalin expression in adult human terminal ileum suggests megalin‐independent cubilin/amnionless activity during vitamin B12 absorption. Physiol Rep 2014;2:e12086. [Crossref] [PubMed]
- Goodman BE. Insights into digestion and absorption of major nutrients in humans. Adv Physiol Educ 2010;34:44-53. [Crossref] [PubMed]
- Li N, Wang D, Sui Z, et al. Development of an improved three-dimensional in vitro intestinal mucosa model for drug absorption evaluation. Tissue Eng Part C Methods 2013;19:708-19. [Crossref] [PubMed]
- Festen H. Intrinsic factor secretion and cobalamin absorption: physiology and pathophysiology in the gastrointestinal tract. Scand J Gastroenterol 1991;26:1-7. [Crossref] [PubMed]
- Hofmann AF, Hagey LR, Krasowski MD. Bile salts of vertebrates: structural variation and possible evolutionary significance. J Lipid Res 2010;51:226-46. [Crossref] [PubMed]
- Leal J, Smyth HDC, Ghosh D. Physicochemical properties of mucus and their impact on transmucosal drug delivery. Int J Pharm 2017;532:555-72. [Crossref] [PubMed]
- Bansil R, Turner BS. Mucin structure, aggregation, physiological functions and biomedical applications. Curr Opin Colloid Interface Sci 2006;11:164-70. [Crossref]
- Prasanna LC. Analysis of the Distribution of Mucins in Adult Human Gastric Mucosa and Its Functional Significance. J Clin Diagn Res 2016;10:AC01-4. [Crossref] [PubMed]
- Behera SK, Praharaj AB, Dehury B, et al. Exploring the role and diversity of mucins in health and disease with special insight into non-communicable diseases. Glycoconj J 2015;32:575-613. [Crossref] [PubMed]
- McGuckin MA, Lindén SK, Sutton P, et al. Mucin dynamics and enteric pathogens. Nat Rev Microbiol 2011;9:265-78. [Crossref] [PubMed]
- Thornton DJ, Rousseau K, McGuckin MA. Structure and function of the polymeric mucins in airways mucus. Annu Rev Physiol 2008;70:459-86. [Crossref] [PubMed]
- Hasnain SZ, Dawson PA, Lourie R, et al. Immune-driven alterations in mucin sulphation is an important mediator of Trichuris muris helminth expulsion. PLoS Pathog 2017;13:e1006218. [Crossref] [PubMed]
- Perez-Vilar J, Hill RL. The structure and assembly of secreted mucins. J Biol Chem 1999;274:31751-4. [Crossref] [PubMed]
- Van den Steen P, Rudd PM, Dwek RA, et al. Concepts and principles of O-linked glycosylation. Crit Rev Biochem Mol Biol 1998;33:151-208. [Crossref] [PubMed]
- Thanka Christlet TH, Veluraja K. Database analysis of O-glycosylation sites in proteins. Biophys J 2001;80:952-60. [Crossref] [PubMed]
- Thanka Christlet TH, Veluraja K. Database analysis of O-glycosylation sites in proteins. Biophys J 2001;80:952-60. [Crossref] [PubMed]
- Zhu L, Lee P, Yu D, et al. Cloning and characterization of human MUC19 gene. Am J Respir Cell Mol Biol 2011;45:348-58. [Crossref] [PubMed]
- Corfield AP. Mucins: a biologically relevant glycan barrier in mucosal protection. Biochim Biophys Acta 2015;1850:236-52. [Crossref] [PubMed]
- Thai P, Loukoianov A, Wachi S, et al. Regulation of airway mucin gene expression. Annu Rev Physiol 2008;70:405-29. [Crossref] [PubMed]
- Ambort D, Johansson ME, Gustafsson JK, et al. Calcium and pH-dependent packing and release of the gel-forming MUC2 mucin. Proc Natl Acad Sci U S A 2012;109:5645-50. [Crossref] [PubMed]
- Ambort D, van der Post S, Johansson ME, et al. Function of the CysD domain of the gel-forming MUC2 mucin. Biochem J 2011;436:61-70. [Crossref] [PubMed]
- Axelsson MA, Asker N, Hansson GC. O-glycosylated MUC2 monomer and dimer from LS 174T cells are water-soluble, whereas larger MUC2 species formed early during biosynthesis are insoluble and contain nonreducible intermolecular bonds. J Biol Chem 1998;273:18864-70. [Crossref] [PubMed]
- Gum JR Jr, Hicks JW, Toribara NW, et al. Molecular cloning of human intestinal mucin (MUC2) cDNA. Identification of the amino terminus and overall sequence similarity to prepro-von Willebrand factor. J Biol Chem 1994;269:2440-6. [Crossref] [PubMed]
- Johansson ME, Larsson JM, Hansson GC. The two mucus layers of colon are organized by the MUC2 mucin, whereas the outer layer is a legislator of host-microbial interactions. Proc Natl Acad Sci U S A 2011;108:4659-65. [Crossref] [PubMed]
- Johansson ME, Phillipson M, Petersson J, et al. The inner of the two Muc2 mucin-dependent mucus layers in colon is devoid of bacteria. Proc Natl Acad Sci U S A 2008;105:15064-9. [Crossref] [PubMed]
- Allen A, Bell A, Mantle M, et al. The structure and physiology of gastrointestinal mucus. Adv Exp Med Biol 1982;144:115-33. [Crossref] [PubMed]
- Wilcox MD, Van Rooij LK, Chater PI, et al. The effect of nanoparticle permeation on the bulk rheological properties of mucus from the small intestine. Eur J Pharm Biopharm 2015;96:484-7. [Crossref] [PubMed]
- Rousseau K, Swallow DM. Mucin methods: genes encoding mucins and their genetic variation with a focus on gel-forming mucins. Methods Mol Biol 2012;842:1-26. [Crossref] [PubMed]
- Groo AC, Lagarce F. Mucus models to evaluate nanomedicines for diffusion. Drug Discov Today 2014;19:1097-108. [Crossref] [PubMed]
- Lock JY, Carlson TL, Carrier RL. Mucus models to evaluate the diffusion of drugs and particles. Adv Drug Deliv Rev 2018;124:34-49. [Crossref] [PubMed]
- Chen Y, Lin Y, Davis KM, et al. Robust bioengineered 3D functional human intestinal epithelium. Sci Rep 2015;5:13708. [Crossref] [PubMed]
- Araújo F, Sarmento B. Towards the characterization of an in vitro triple co-culture intestine cell model for permeability studies. Int J Pharm 2013;458:128-34. [Crossref] [PubMed]
- Foster SN, Pearson JP, Hutton DA, et al. Interaction of polyacrylates with porcine pepsin and the gastric mucus barrier: a mechanism for mucosal protection. Clin Sci (Lond) 1994;87:719-26. [Crossref] [PubMed]
- Friedl H, Dünnhaupt S, Hintzen F, et al. Development and evaluation of a novel mucus diffusion test system approved by self-nanoemulsifying drug delivery systems. J Pharm Sci 2013;102:4406-13. [Crossref] [PubMed]
- Bhat PG, Flanagan DR, Donovan MD. Drug diffusion through cystic fibrotic mucus: steady-state permeation, rheologic properties, and glycoprotein morphology. J Pharm Sci 1996;85:624-30. [Crossref] [PubMed]
- Norris DA, Sinko PJ. Effect of size, surface charge, and hydrophobicity on the translocation of polystyrene microspheres through gastrointestinal mucin. J Appl Polym Sci 1998;63:1481-92. [Crossref]
Cite this article as: Stanforth K, Chater P, Brownlee I, Wilcox M, Ward C, Pearson J. In vitro modelling of the mucosa of the oesophagus and upper digestive tract: narrative review. Ann Esophagus 2022;5:4.


