
Molecular effects of tobacco smoke and bile reflux in Barrett’s esophagus in vitro
Introduction
Esophageal adenocarcinoma (EAC) has become one of the most rapidly increasing cancers among Americans in the last decade, with the disease having one of the highest mortality rates among all forms of cancer (1). The chronic condition known as gastroesophageal reflux disease creates an environment of high oxidative stress in the esophagus that is believed to contribute to the carcinogenic transformation from reflux esophagitis to the premalignant condition known as Barrett’s esophagus (BE), and then ultimately to EAC (2).
Tobacco smoke has been significantly implicated in a number of cancers including lung, oral, head and neck, and esophageal cancers (3). Although smoking has been shown to cause both squamous cell carcinoma and adenocarcinoma of the esophagus, the risk of EAC remains relatively constant among smokers and ex-smokers (4). Only recently has the association of cigarette smoking and EAC been described (5-11). The mechanisms through which tobacco smoke may lead to malignant transformation of the esophagus include: (I) a dose-dependent increase in acid secretion by the gastric parietal cells; (II) increased levels of duodenogastric reflux causing increased concentrations of bile acid in the refluxate; (III) relaxation of the lower esophageal sphincter; (IV) decreased secretion of protective mucus; and (V) induced histamine-mediated susceptibility to mucosal damage (12). We have recently demonstrated molecular changes in the esophageal epithelium after a subchronic exposure to cigarette smoke in the presence of bile-acid reflux (13). In this study we demonstrated that cigarette smoke aggravates reflux-induced BE and could potentially accelerate the progression of BE to EAC through the loss of MnSOD, and over-expression of NF kappa B and COX-2 mediated factors. However, the specific changes of molecular events linked to carcinogenesis markers have not yet been elucidated.
The transcription factor nuclear factor-κB (NF-κB) is activated in esophageal epithelial cells by bile acid and acidic pH in an experimental animal model of GERD (14,15), and in BE patients (16). Cigarette smoke extract activates NF-κB via the NF-κB-inducing kinase/IκB kinase (NIK/IKK) pathway in various cell types (17). However, these studies have been mostly performed in tissues and cell types other than esophageal. The effects of tobacco smoke and its constituents on the molecular pathways in the pathologies of the esophagus, especially in the presence of reflux and/or bile acids, remain largely unknown.
There have been a wide range of genes that have been implicated as potentially having a role in the development of BE and EAC, including MUC, CDX2, COX2, SOD2, and other genes. The MUC genes are responsible for protecting the epithelial lining of the esophagus. The CDX2 gene is physiologically expressed throughout the small and large intestines and is responsible for the differentiation and maintenance of the simple columnar epithelium of the intestines (18). Expression of CDX2 in the esophageal epithelium has been linked to the development of esophageal metaplasia and BE; therefore, CDX2 is believed to be an early marker for dysplastic epithelium of the esophagus (18). Previous experimental studies have shown significant over-expression of the COX2 gene in patients diagnosed with BE and EAC (2). However, no one has evaluated how these genes are affected by bile acid and smoke exposure in combination, as would occur when a patient with gastroesophageal reflux disease (GERD) smokes.
In this in vitro study, we aimed to determine the effects of standardized smoke condensate and bile acid treatments on many of these genes that have been linked to the development of BE and EAC.
Methods
Cell culture and treatment
The two cell lines evaluated in this study were the human hTERT-immortalized non-neoplastic Barrett’s esophageal cell line (BarT) and the human esophageal squamous cell line (Het-1a) as previously reported (2,19). The cell viability was not changed across treatments as we have previously published (13).
Before experimental treatment of the cell lines, standardized bile acid mixtures consisting of equimolar concentrations of sodium taurocholate, sodium glycocholate, sodium cholate, and sodium deoxycholate in both the Het-1a and BarT media were prepared at concentrations of 0.8 mM. In addition, Het-1a and BarT cell line media were conditioned with cigarette smoke from 2R1 Kentucky reference cigarettes (Kentucky Tobacco Research Center, Lexington, KY) according to method described (20) and was performed in Dr. David A. Scott’s laboratory at the University of Louisville. The nicotine equivalents in the stock media were 8,000 ng/mL for Het-1a and BarT cell in their respective media. The nicotine measurements were done at the UCSF-Clinical Pharmacology Laboratory, San Francisco, CA.
When four 75 mL flasks of the BarT cells or Het-1a cells had grown to approximately 80% confluence, the cells were transferred into four six-well plates at approximately 0.5×106 cells per well. The cells in the six-well plates were then treated via the experimental protocol. Each treatment regimen used three wells of one of the six-well plates. The experimental control was designated as the “Untreated Control,” and each well for the control received 1.5 mL of cell medium at the beginning of the treatment protocol. The first regimen was designated as the “4,000 ng Smoke” treatment, and each well for this regimen received 1.5 mL mixtures of regular cell medium and cigarette smoke-conditioned cell medium so that the final nicotine concentration was 4,000 ng/mL. The second treatment regimen was designated as the “0.4 mM Bile” treatment, and each well in these regimens received 1.5 mL mixtures of regular cell medium and standardized bile acid mixture in cell medium so that the final bile acid concentration was 0.4 mM. The third treatment regimen received 1.5 mL mixtures of the cigarette smoke-conditioned cell medium and the standardized bile acid mixture in cell medium per well. These treatments were designated as “Combination” and received 1.5 mL mixtures of cigarette smoke and bile acid conditioned media so that the final concentrations were 4,000 ng/mL smoke condensate and 0.4 mM bile acid. Based on our pilot results of cell tolerance to the bile and smoke challenge, Het-1a cells were treated for 4 hours, while BarT cells for 4 and 24 hours. After the treatments, the six-well plates of cells were placed back in the incubator at 37 °C in a humidified atmosphere of 5% CO2 for 4 hours before RNA was extracted. The identical protocol was repeated for 24-hour treatment, after which RNA was extracted.
Reverse transcription of RNA to cDNA
The RNA extraction was performed via standard technique and the RNA samples were removed from the −80 °C freezer and allowed to thaw. Based on their RNA concentrations, the samples were diluted with the appropriate volume of nuclease-free water so that the RNA concentration of each sample was 40 ng/µL. Then, 25 µL of each RNA sample and 25 µL of Reverse Transcription Master Mix (Applied Biosystems; Foster City, CA) prepared by the manufacturer’s instructions were added to individual 200 µL tubes. The Applied Biosystems 2720 Thermal Cycler was then used to reverse transcribe the mRNA into cDNA. The cDNA was then diluted to 150 µL of nuclease-free water so that the final cDNA concentration for each sample was 6.67 ng/µL.
Relative quantification real-time PCR
Relative gene expression levels were measured via relative quantification real-time PCR in the Applied Biosystems 7300 Real Time PCR System using the SYBR Green method (Applied Biosystems). Each cDNA sample (3 µL; 20 ng) was run in triplicate for each gene of interest on 96-well PCR plates. β-actin was used as the reference gene for comparison of the cycle threshold (CT) values. The mean CT value for β-actin among all treatment regimens in the Het-1a cell line samples and the mean CT value for β-actin among all treatment regimens in the BarT cell line samples were used for comparison to CT values for all other genes. The standard 2-ΔΔCt method for relative quantification real-time PCR was utilized to determine the gene expression for each sample relative to the gene expression for the Untreated Control samples using the mean CT value for β-actin as reference.
Statistical analyses
Statistical analyses were performed via two-tailed unpaired student t-tests which were used to evaluate the variance with respect to the fold change values of gene expression among the various treatment regimens for each gene in the BarT 4-hour, BarT 24-hour, and Het-1a 4-hour groups. Results of the statistical analyses are reported as probability values (P values) with those values ≤0.05 considered significant.
Results
Relative quantification real-time PCR was utilized to determine the relative changes in gene expression for fourteen different genes in the BarT cell line and Het-1a cell line following standardized bile acid and smoke condensate treatment regimens.
Figure 1 shows the average relative fold changes of the four studied MUC genes after treatment relative to the untreated control. MUC1 was not detectable in the BarT cell line treated for 4 hours. MUC2 was detectable in the Het-1a cells in the bile acid treatment alone group. P values are reported on the figure.
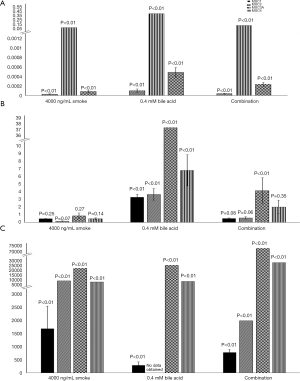
Figure 2 illustrates the average relative fold changes of the three studied genes pertinent to oxidative stress, SOD1, SOD2, and COX2 after the different treatments in the different groups. P values are reported on the figure.
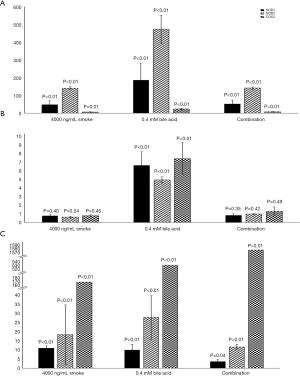
Figure 3 shows the average relative fold changes of the five studied NF-kB regulated genes. All five genes were detectable in the BarT 4 hour cells, but TNF-Alpha, IFN-Gamma and IL-1Beta data showed no significant changes in the BarT 24-hour cells. TNF-alpha was not detectable in the Het-1a cells exposed to smoke alone or bile alone. TNF-alpha, IFN-Gamma, IL-1Beta, and IL-6 were not detectable in the Het-1a cells exposed to bile alone. All five genes were detectable in the Het-1a cells exposed to a combination of smoke and bile. P values are reported on the figure.
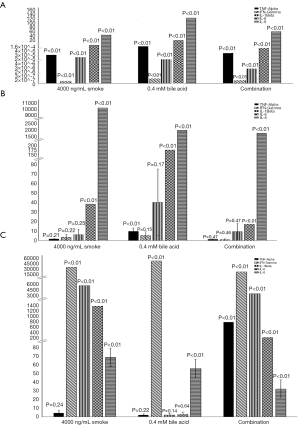
Figure 4 displays the average relative fold change in CDX2 expression from the different treatments in all three cell line groups. P values are reported on the figure.
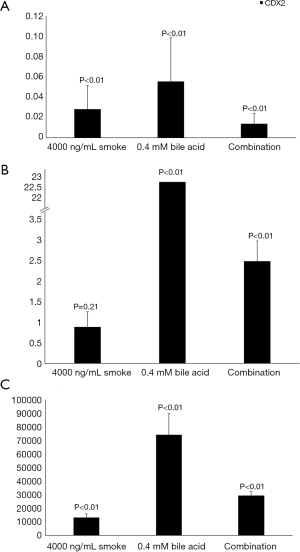
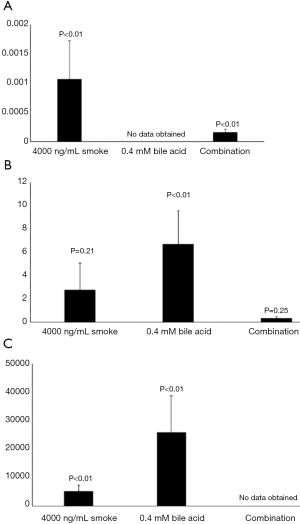
Figure 5 shows the average relative fold change in CYP3A4 expression from the different treatments in all three cell groups. It was not detectable for bile acid alone treatment in the BarT cell line exposed for 4 hours. It was not detectable for combination treatment in the Het-1a cells. P values are reported on the figure.
Discussion
This study aimed to determine the effects of different tobacco smoke condensate and bile acid treatments in a Barrett’s esophageal cell line (BarT) and a normal esophageal squamous cell line (Het-1a) on the expression of various genes that may be involved in the development and carcinogenic progression of BE. Our previous studies have demonstrated the effects of a combination of reflux and cigarette smoke in animal models (13). Combination of duodenogastroesophageal reflux and cigarette smoke in in vivo models has been shown to activate NF-κB and significantly increase expression of the COX2 gene (13). The purpose of this study was to perform in vitro confirmation of many of the trends observed in the in vivo study and explore other molecular effects of bile acid refluxate and tobacco smoke on the esophageal epithelium.
The MUC genes are responsible for protecting the epithelial lining of the esophagus by coding for high molecular weight glycoproteins known as mucins. In this study, four MUC genes were evaluated to determine the effect of tobacco smoke and bile acids on their expression. The BarT cells after 4 hours of all three treatments showed a varying degree of heavy downregulation in the MUC gene family, suggesting a loss of the protective function of these glycoproteins in the precancerous condition. The 24-hour treatment of this cell line, however, displays an insignificant change in MUC expression from combination or smoke treatment alone. This group showed an upregulation in MUC expression from bile acid exposure alone. The differences in relative transcription suggests that time of exposure may play a role in gene expression when these cells are subjected to smoke, refluxate or both. The Het-1a cells treated with smoke, bile acid, or a combination revealed an enormous upregulation across all four MUC genes studied from all treatments. These results show that the insult of smoke condensate, bile acid refluxate, or a combination of the two invokes a response that increases their mRNA levels.
Analysis of the genes related to oxidative stress in the BarT cell line and Het-1a cell line following the smoke condensate and bile acid treatments revealed some interesting patterns. The BarT cells treated for 4 hours experienced a sharp upregulation in both superoxide dismutase genes, and an upregulation of COX2, though to a smaller degree. The 24-hour exposure of this cell line to smoke condensate alone, interestingly, caused a downregulation of all three genes, highlighting the possibility that time of exposure may change the transcriptional response. The 0.4 mM bile acid exposure for 24 hours caused an upregulation of all three genes. The combination treatment for 24 hours in BarT cells caused a slight downregulation of SOD1, very little change in SOD2 expression, and a slight increase in COX2 expression. All treatments to the Het-1a cells for 4 hours caused an increase in all three genes, to varying degrees. The trend in COX2 expression from the combination treatment in two cell lines is of particular interest due to its prominent role as an inflammatory mediator and potential role in carcinogenic processes, implying a blunt response of BarT cells compared to Het-1a.
In the current study, activation of the NFκB was measured indirectly by analyzing the changes in expression of five of the genes that are regulated by NF-κB: TNFα, IFNγ, IL1β, IL6, and IL8. The results of this study confirm that, with the exception of TNF-α, IFN-gamma, and IL-1β in the BarT 4-hour cells, NF-κB-regulated genes are synergistically activated by treatment regardless of the cell line (21). Interestingly, these findings largely coincide with in vivo reports that showed increased expression of NF-κB inducing kinase in the esophageal tissue of esophageal-duodenal anastomosis rat models following controlled cigarette smoke administration (13). Future work must be done to determine the precise mechanism in which NF-κB inducing kinase and the NF-κB regulated genes in the esophageal mucosa are affected by tobacco smoke and refluxate. This activation of NF-κB in these cell lines is of particular interest due to the implications of aberrant NF-κB expression in different cancers, and these results suggest a potential route of molecular carcinogenesis.
The CDX2 gene plays a very significant role in the development of BE because of its role in regulating differentiation and maintenance of intestinal simple columnar epithelia. After 4 hours of exposure to any of the treatments, the BarT cells experienced sharp downregulation of CDX2. The 24-hour exposure of smoke condensate alone to this cell line caused an insignificant change in CDX2 expression, though bile acid and combination treatment caused varying degrees of upregulation. The Het-1a cells exposed to any of the treatments for 3 hours showed a very sharp increase in CDX2 expression to varying degrees. These results highlight potential functional differences between precancerous lesions encountered in BE and normal esophageal epithelium.
The CYP3A4 gene belongs to the cytochrome p450 superfamily of enzymes that play an important role in toxic compound metabolism. Down-regulation of CYP3A4 has been implicated in the carcinogenic progression of BE by previous study (22). The 4-hour exposure of smoke condensate alone and combination treatment caused a sharp decrease in CYP3A4 expression in BarT cells. 24-hour exposure of smoke condensate alone and bile acid alone to this cell line caused an increase in CYP3A4 expression, but interestingly the combination treatment for 24 hours caused a decrease in CYP3A4 expression. The Het-1a cell lines exposed for 4 hours revealed a large upregulation of CYP3A4 from smoke condensate alone and bile acid alone. These results highlight the difference in response to cigarette smoke and bile acid in the epithelium of the precancerous condition compared to normal esophageal epithelium.
Although this study provides valuable insights into how many of the genes commonly associated with the development of EAC may be affected by duodenogastroesophageal reflux and tobacco smoking, there are some limits to these results that are at mRNA levels only. Nevertheless, the in vitro data from this study has revealed some readily observable patterns of altered gene expression, many of which are similar to those observed in the previous in vivo studies. These include up-regulation of some of the NF-κB-regulated genes, namely the interleukins, by combination smoke condensate and bile acid treatments as well as dysregulation of SOD1, SOD2 and COX2 by combination smoke condensate and bile acid treatment in both the BarT cell line and Het-1a cell line. Additionally, this study provided novel information regarding the effects of tobacco smoke and bile acids on the MUC genes responsible for protecting the esophageal epithelium, revealing significant down-regulation by combination treatments in the BarT cell lines and an upregulation of the mucin family in Het-1a cells, perhaps hinting at a difference in the precancerous tissue vs. normal esophageal epithelium. Therefore, this study provides further evidence for a synergistic relationship between tobacco smoke and acid reflux in altering expression of many genes commonly associated with the carcinogenic progression of BE to a greater extent in combination than alone. Smoking is still commonly viewed as a secondary risk factor for the development of EAC because of the physiological changes it induces that lead to exposure of the esophageal mucosa to increased refluxate. However, our study suggests that smoking may in fact be a primary risk factor for the development of EAC because of the direct effects that tobacco smoke, when combined with refluxate, has on gene expression in the esophageal mucosa.
Acknowledgments
Funding: None.
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/aoe.2019.09.01). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). Institutional ethical approval and individual informed consent were waived.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Jemal A, Bray F, Center MM, et al. Global cancer statistics. CA Cancer J Clin 2011;61:69-90. [Crossref] [PubMed]
- Schiffman SC, Li Y, Xiao D, et al. The resistance of esophageal adenocarcinoma to bile salt insult is associated with manganese superoxide dismutase expression. J Surg Res 2011;171:623-30. [Crossref] [PubMed]
- DeMarini DM. Genotoxicity of tobacco smoke and tobacco smoke condensate: a review. Mutat Res 2004;567:447-74. [Crossref] [PubMed]
- Bosetti C, Gallus S, Garavello W, et al. Smoking cessation and the risk of oesophageal cancer: An overview of published studies. Oral Oncol 2006;42:957-64. [Crossref] [PubMed]
- Casson AG, Zheng Z, Porter GA, et al. Genetic polymorphisms of microsomal epoxide hydroxylase and glutathione S-transferases M1, T1 and P1, interactions with smoking, and risk for esophageal (Barrett) adenocarcinoma. Cancer Detect Prev 2006;30:423-31. [Crossref] [PubMed]
- Cook MB, Shaheen NJ, Anderson LA, et al. Cigarette smoking increases risk of Barrett's esophagus: an analysis of the Barrett's and Esophageal Adenocarcinoma Consortium. Gastroenterology 2012;142:744-53. [Crossref] [PubMed]
- Lubin JH, Cook MB, Pandeya N, et al. The importance of exposure rate on odds ratios by cigarette smoking and alcohol consumption for esophageal adenocarcinoma and squamous cell carcinoma in the Barrett's Esophagus and Esophageal Adenocarcinoma Consortium. Cancer Epidemiol 2012;36:306-16. [Crossref] [PubMed]
- Tramacere I, La Vecchia C, Negri E. Tobacco smoking and esophageal and gastric cardia adenocarcinoma: a meta-analysis. Epidemiology 2011;22:344-9. [Crossref] [PubMed]
- Yoon HH, Lewis MA, Shi Q, et al. Prognostic impact of body mass index stratified by smoking status in patients with esophageal adenocarcinoma. J Clin Oncol 2011;29:4561-7. [Crossref] [PubMed]
- Zhai R, Chen F, Liu G, et al. Interactions among genetic variants in apoptosis pathway genes, reflux symptoms, body mass index, and smoking indicate two distinct etiologic patterns of esophageal adenocarcinoma. J Clin Oncol 2010;28:2445-51. [Crossref] [PubMed]
- Zhai R, Liu G, Asomaning K, et al. Genetic polymorphisms of VEGF, interactions with cigarette smoking exposure and esophageal adenocarcinoma risk. Carcinogenesis 2008;29:2330-4. [Crossref] [PubMed]
- Endoh K, Leung FW. Effects of smoking and nicotine on the gastric mucosa: a review of clinical and experimental evidence. Gastroenterology 1994;107:864-78. [Crossref] [PubMed]
- Aiyer HS, Li Y, Harper N, et al. Molecular changes in the esophageal epithelium after a subchronic exposure to cigarette smoke in the presence of bile-acid reflux. Inhal Toxicol 2011;23:304-11. [Crossref] [PubMed]
- Lee JS, Oh TY, Ahn BO, et al. Involvement of oxidative stress in experimentally induced reflux esophagitis and Barrett's esophagus: clue for the chemoprevention of esophageal carcinoma by antioxidants. Mutat Res 2001;480-481:189-200. [Crossref] [PubMed]
- Abdel-Latif MM, Inoue H, Kelleher D, et al. Factors regulating nuclear factor-kappa B activation in esophageal cancer cells: Role of bile acids and acid. J Cancer Res Ther 2016;12:364-73. [Crossref] [PubMed]
- Jenkins GJ, Harries K, Doak SH, et al. The bile acid deoxycholic acid (DCA) at neutral pH activates NF-kappaB and induces IL-8 expression in oesophageal cells in vitro. Carcinogenesis 2004;25:317-23. [Crossref] [PubMed]
- Kim SR, Jung YR, Kim DH, et al. Caffeic acid regulates LPS-induced NF-kappaB activation through NIK/IKK and c-Src/ERK signaling pathways in endothelial cells. Arch Pharm Res 2014;37:539-47. [Crossref] [PubMed]
- Weimann A, Rieger A, Zimmermann M, et al. Comparison of six immunohistochemical markers for the histologic diagnosis of neoplasia in Barrett's esophagus. Virchows Arch 2010;457:537-45. [Crossref] [PubMed]
- Schiffman SC, Li Y, Martin RC. The Association of Manganese Superoxide Dismutase Expression in Barrett's Esophageal Progression With MnTBAP and Curcumin Oil Therapy. J Surg Res 2012;176:535-41. [Crossref] [PubMed]
- Bagaitkar J, Williams LR, Renaud DE, et al. Tobacco-induced alterations to Porphyromonas gingivalis-host interactions. Environ Microbiol 2009;11:1242-53. [Crossref] [PubMed]
- Fitzgerald RC, Onwuegbusi BA, Bajaj-Elliott M, et al. Diversity in the oesophageal phenotypic response to gastro-oesophageal reflux: immunological determinants. Gut 2002;50:451-9. [Crossref] [PubMed]
- Hughes SJ, Morse MA, Weghorst CM, et al. Cytochromes P450 are expressed in proliferating cells in Barrett's metaplasia. Neoplasia 1999;1:145-53. [Crossref] [PubMed]
Cite this article as: Pulliam Z, Harper N, Li Y, Farmer R, Martin RCG. Molecular effects of tobacco smoke and bile reflux in Barrett’s esophagus in vitro. Ann Esophagus 2019;2:17.


